2425-m1-geniomhe-group-6
Lab 3 Report
Table of contents
- Lab 3 Report
First Implementation
This implementation is in the branch previous-model-extension .
In this lab, we extended our previous model to include the following functionalities:
- The parser returns a numpy array representation of the molecule.
- Writing structures into PDML/XML format.
First, we kept our previous model and extended it with the minimal changes possible without using the design patterns.
Class Diagram for the Previous-Model-Extension
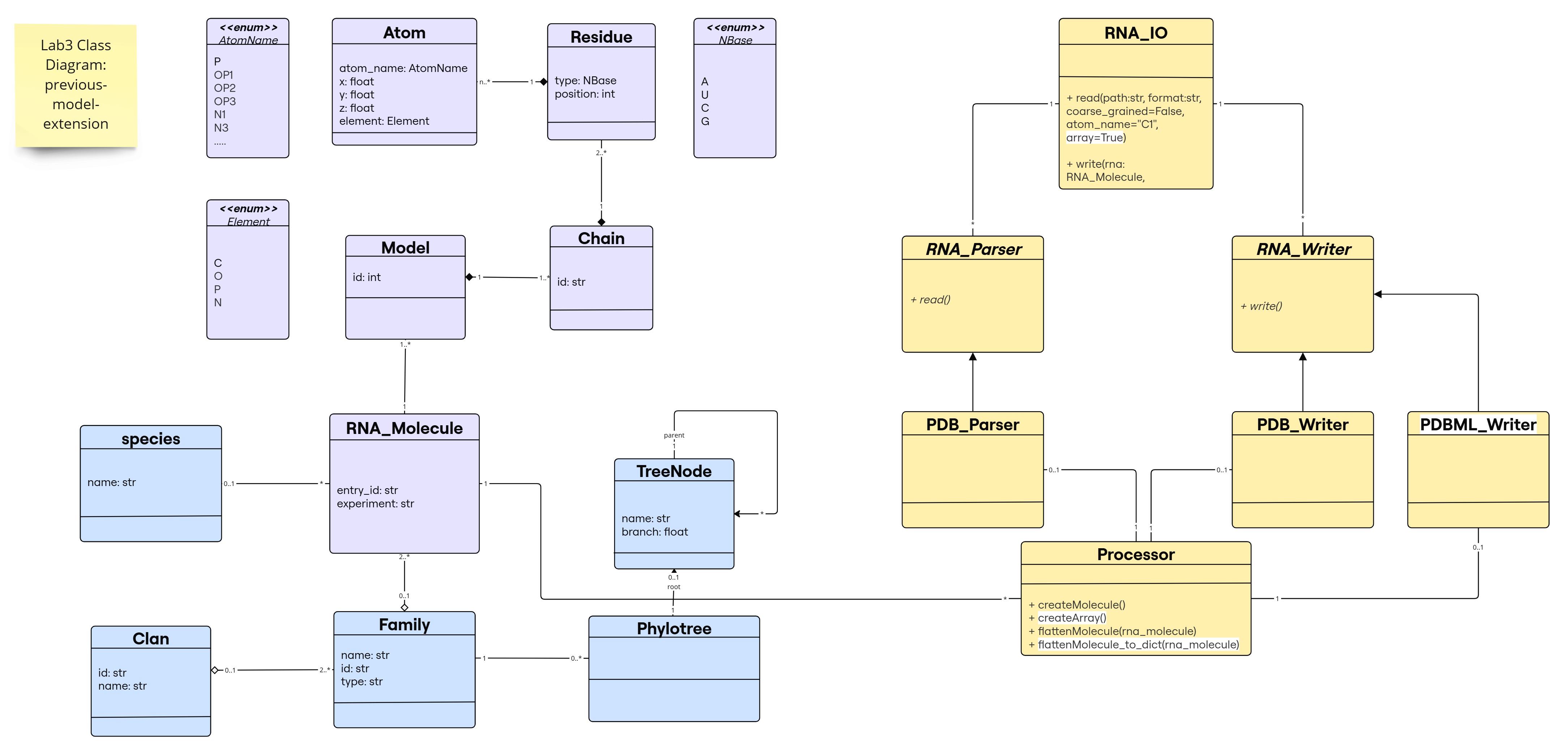
The new changes are highlighted in white; note that flattenMolecule_to_dict() is a new method to provide an extra utility to the Processor class, not mandatory for the implementation (e.g., some libraries like numpy, pandas, etc. provide similar functionalities that transforms their main objects to different types of data structures).
Object Diagram for the Previous-Model-Extension
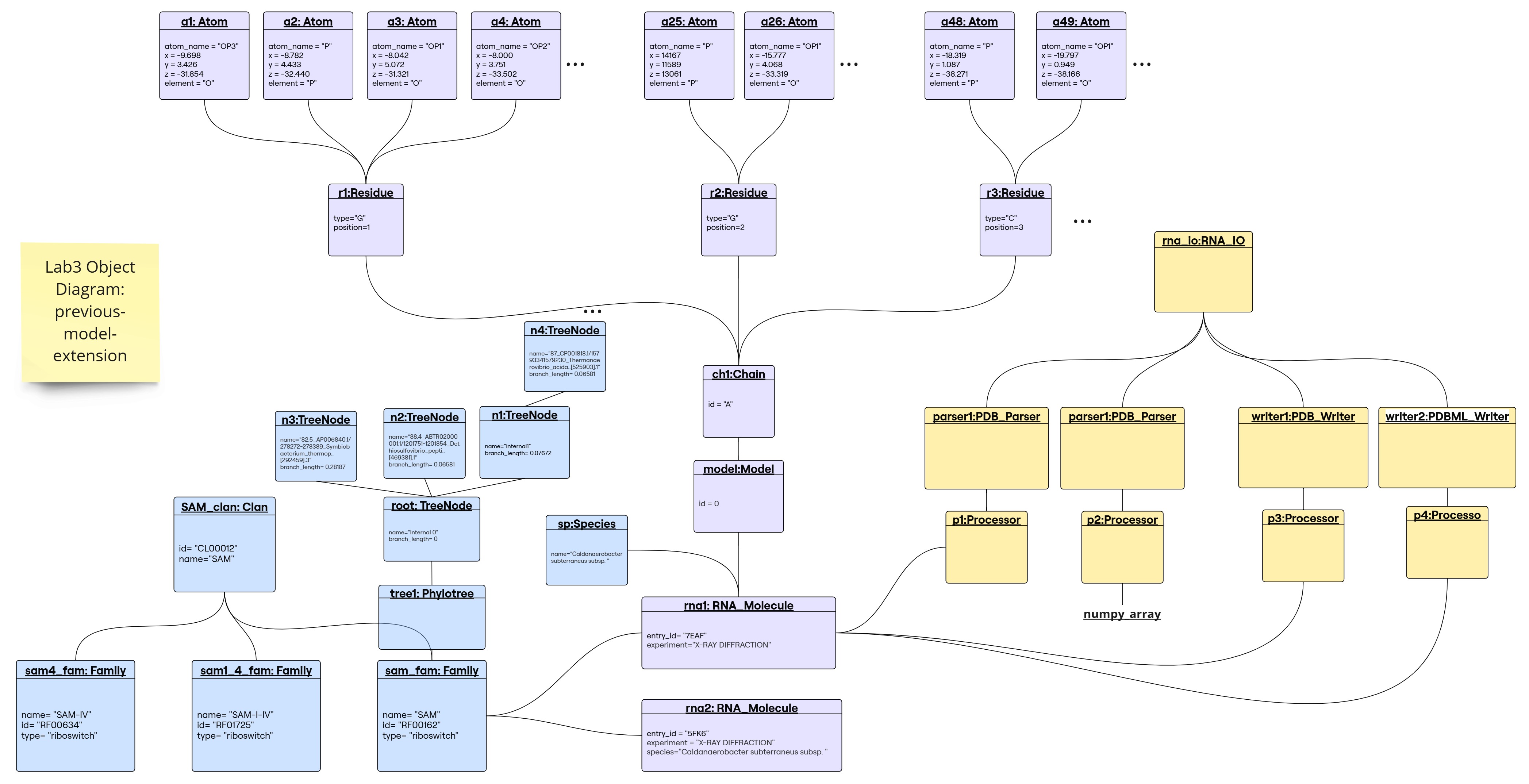
1. Parser returns a numpy array
For this, we added a function in the Processor class called: createArray() that returns the required numpy array representation of an RNA Molecule that can have multiple models.
Note: In our previous implementation, the parser will store the atom information in a list atoms inside the Processor class.
createArray() in Processor class:
- Returns a numpy array representation of the molecule
- Dimension:
(number of models, max_residues_no, max_atoms_per_residue_no, 3) - Stores the x,y,z coordinates of each atom.
- Implementation notes:
- To find the number of models we checked the model_id of the last atom in the atoms list.
- We had to loop through the list of atoms to find the maximum number of residues among all models and the maximum number of atoms per residue.
- Considering that the same atom in a residue can have different coordinates (alternate locations), we stored the coordinates of the atom that has the highest occupancy.
- The array is filled with nan values where there is no atom information.
read() in PDB_Parser class:
We added a boolean argument array to the read() function and set it to True by default. If the argument is True, the function will return the numpy array representation of the molecule, otherwise it will create the molecule object as before. We did not change anything in the read() function, we just added the following at the end:
if array:
return processor.createArray()
else:
return processor.createMolecule()
Code Usage
An example can be found in the notebook reading.ipynb from the other branch.
We read a molecule that contains 1 model and another molecule that contains multiple models, and showed the resulting arrays. A brief example:
rna_io=RNA_IO()
pdb_path_test=pathify_pdb("7eaf")
mol=rna_io.read(pdb_path_test, "PDB")
print(mol.shape)
print(mol[0, -1, 0, :])
The output:
(1, 94, 24, 3)
[-10.06 7.177 -49.234]
2. Writing Structures into PDML/XML format
File format description:
An PDBML file is an XML file that contains protein/nucleic acid structure information within an xml format. This is an efficient data storing format is widely used in databases and software tools to store and exchange data files in a structured manner. It was introduced to PDBe as the “PDBML” format by Westbrook et al. in a 2005 in a paper published in Bioinformatics entitled “PDBML: the representation of archival macromolecular structure data in XML”1.
In python, writing and handling xml files is done here through 2 modules in our code: xml.etree.ElementTree and xml.dom.minidom. The first module is used to create the xml file and the second module is used to prettify the xml file (provide proper xml indentation).
Starting off by exploring this file (example used is 7eaf.xml taken from pdb), we notice the following structure
<PDBx:datablock xmlns:PDBx="http://pdbml.pdb.org/schema/pdbx-v50.xsd" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" datablockName="7EAF" xsi:schemaLocation="http://pdbml.pdb.org/schema/pdbx-v50.xsd pdbx-v50.xsd">
...
</PDBx:datablock>
with all these tags included in the file:
grep -e '^ <PDBx:' 7eaf.xml #to retrive the list
<PDBx:atom_siteCategory><PDBx:atom_site_anisotropCategory><PDBx:atom_sitesCategory><PDBx:atom_typeCategory><PDBx:audit_authorCategory><PDBx:audit_conformCategory><PDBx:cellCategory><PDBx:chem_compCategory><PDBx:chem_comp_atomCategory><PDBx:chem_comp_bondCategory><PDBx:citationCategory><PDBx:citation_authorCategory><PDBx:database_2Category><PDBx:diffrnCategory><PDBx:diffrn_detectorCategory><PDBx:diffrn_radiationCategory><PDBx:diffrn_radiation_wavelengthCategory><PDBx:diffrn_sourceCategory><PDBx:entityCategory><PDBx:entity_polyCategory><PDBx:entity_poly_seqCategory><PDBx:entryCategory><PDBx:exptlCategory><PDBx:exptl_crystalCategory><PDBx:exptl_crystal_growCategory><PDBx:ndb_struct_conf_naCategory><PDBx:ndb_struct_na_base_pairCategory><PDBx:ndb_struct_na_base_pair_stepCategory><PDBx:pdbx_audit_revision_categoryCategory><PDBx:pdbx_audit_revision_detailsCategory><PDBx:pdbx_audit_revision_groupCategory><PDBx:pdbx_audit_revision_historyCategory><PDBx:pdbx_audit_revision_itemCategory><PDBx:pdbx_audit_supportCategory><PDBx:pdbx_database_statusCategory><PDBx:pdbx_entity_nonpolyCategory><PDBx:pdbx_entity_src_synCategory><PDBx:pdbx_entry_detailsCategory><PDBx:pdbx_initial_refinement_modelCategory><PDBx:pdbx_nonpoly_schemeCategory><PDBx:pdbx_poly_seq_schemeCategory><PDBx:pdbx_refine_tlsCategory><PDBx:pdbx_refine_tls_groupCategory><PDBx:pdbx_struct_assemblyCategory><PDBx:pdbx_struct_assembly_auth_evidenceCategory><PDBx:pdbx_struct_assembly_genCategory><PDBx:pdbx_struct_assembly_propCategory><PDBx:pdbx_struct_conn_angleCategory><PDBx:pdbx_struct_oper_listCategory><PDBx:pdbx_struct_special_symmetryCategory><PDBx:pdbx_validate_close_contactCategory><PDBx:pdbx_validate_rmsd_bondCategory><PDBx:refineCategory><PDBx:refine_histCategory><PDBx:refine_ls_restrCategory><PDBx:refine_ls_shellCategory><PDBx:reflnsCategory><PDBx:reflns_shellCategory><PDBx:softwareCategory><PDBx:space_groupCategory><PDBx:space_group_symopCategory><PDBx:structCategory><PDBx:struct_asymCategory><PDBx:struct_connCategory><PDBx:struct_conn_typeCategory><PDBx:struct_keywordsCategory><PDBx:struct_refCategory><PDBx:struct_ref_seqCategory><PDBx:symmetryCategory>
Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. Bioinformatics, 2005, 21(7):988-992. PubMed:15509603 full text
The <PDBx:atom_siteCategory> tag contains all the information about the atoms in the structure, including all the hierarchical information model > chain > residue > atom. Thus the structure is solely defined by a list of atoms, in this format each will be represented by a tag <PDBx:atom_site>.
<PDBx:datablock xmlns:PDBx="http://pdbml.pdb.org/schema/pdbx-v50.xsd" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" datablockName="7EAF" xsi:schemaLocation="http://pdbml.pdb.org/schema/pdbx-v50.xsd pdbx-v50.xsd">
<PDBx:atom_siteCategory>
<PDBx:atom_site id="1">
...
</PDBx:atom_site>
<PDBx:atom_site id="2">
...
</PDBx:atom_siteCategory>
</PDBx:datablock>
[!IMPORTANT] The atom_siteCategory tag is the only category that reflects the information that we’re capturing in this library, whether thorugh the RNA_Molecule object or the numpy array representation of it. This is the only category that will be included in the xml file. Others include information about bonds, symmetry, experimental setting and other metadata that is not captured in our object.
This is how the hierarchy leading to an atom representation is portrayed in the .xml file.
PDBx:datablock
├── datablockName
├── xsi:schemaLocation
└── PDBx:atom_siteCategory
└── PDBx:atom_site
├── id
├── PDBx:B_iso_or_equiv
├── PDBx:Cartn_x
├── PDBx:Cartn_y
├── PDBx:Cartn_z
├── PDBx:auth_asym_id
├── PDBx:auth_atom_id
├── PDBx:auth_comp_id
├── PDBx:auth_seq_id
├── PDBx:group_PDB
├── PDBx:label_alt_id
├── PDBx:label_asym_id
├── PDBx:label_atom_id
├── PDBx:label_comp_id
├── PDBx:label_entity_id
├── PDBx:label_seq_id
├── PDBx:occupancy
├── PDBx:pdbx_PDB_model_num
└── PDBx:type_symbol
Notice a slight difference between the representation of an atom with $occupancy=1$ and an atom with $occupancy<1$ (having an altrnate location). The difference is the presence of the label_alt_id tag. This will be taken care of while writing the file. This being said, each alternative location of an atom is cosidered a different atom in the file (with PDBx:atom_site id being $+1$ the id of the previous alternate location).
| Atom with no alternative location | Atom with alternative location |
|---|---|
|
|
in this example id 171 is alt location B of the same atom in 170, and shows different occupancy
Implementation: object to xml
Thanks to the hierarchical class design of the molecule object, we’re able to retrieve all information needed describing an atom, for each atom in the molecule.
In porcessor, this method flattenMolecule_to_dict takes an object and returns a list of atom dictionaries, where the keys of each dictionary are named exactly as the tags in the xml file.
[!NOTE] It practically has the same behavior as
flattenMoleculebut returns a list of dictionaries instead of a list of atom info and objects (allowing diverse output formats).
This way, we can easily create the xml file by iterating over the list of atoms and creating the corresponding tags.
def flattenMolecule_to_dict(self,rna_molecule:RNA_Molecule):
'''
rna_molecule: RNA_Molecule object -> RNA molecule to be flattened -> list of atom dictionaries
'''
atoms_list = []
for model_num,_ in enumerate(rna_molecule.get_models()): #--looping through all models
model=rna_molecule.get_models()[_] # --model object from dict key
for chain in model.get_chains().values(): #--looping through all chains
for residue in chain.get_residues().values(): #--looping through all residues
for atom_key, atom in residue.get_atoms().items(): #--looping through all atoms
atom_id, alt_id = atom_key # unpacking atom key (alt_id is '' if no alt location)
# --keys defined identically to pdbml format, values extracted directly from atom object
atom_data = {
"atom_id": str(len(atoms_list) + 1), # Assign a sequential ID
"B": str(atom.temp_factor),
"x": str(atom.x),
"y": str(atom.y),
"z": str(atom.z),
"chain_id": chain.id,
"atom_id": atom_id,
"residue_type": residue.type.name,
"residue_pos": str(residue.position),
"alt_id": None if alt_id == "" else alt_id,
"occupancy": str(atom.occupancy),
"model_no": model_num+1,
"atom_element": atom.element.name
}
atoms_list.append(atom_data)
return atoms_list
To convert to PDBML, xml formatting private functions have been implemented in PDBML_Writer submodule.
# --helper methods
def _wrap_str_to_xml(self,s,name='pdbml_output.xml'):
with open(name, "w") as f:
f.write(s)
def _format_atom_info(self, atoms_list,entry_id):
'''
formats a list of atoms dicts into XML format
'''
s='''<?xml version="1.0" encoding="UTF-8" ?>
<PDBx:datablock datablockName="'''+entry_id+'''"
xmlns:PDBx="http://pdbml.pdb.org/schema/pdbx-v50.xsd"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://pdbml.pdb.org/schema/pdbx-v50.xsd pdbx-v50.xsd">'''
s+='\n\t<PDBx:atom_siteCategory>\n'
for atom in atoms_list:
s+='\t\t<PDBx:atom_site id="'+atom["atom_id"]+'">\n'
s+='\t\t\t<PDBx:B_iso_or_equiv>'+str(atom['B'])+'</PDBx:B_iso_or_equiv>\n'
s+='\t\t\t<PDBx:Cartn_x>'+str(atom['x'])+'</PDBx:Cartn_x>\n'
s+='\t\t\t<PDBx:Cartn_y>'+str(atom['y'])+'</PDBx:Cartn_y>\n'
s+='\t\t\t<PDBx:Cartn_z>'+str(atom['z'])+'</PDBx:Cartn_z>\n'
s+='\t\t\t<PDBx:auth_asym_id>'+atom['chain_id']+'</PDBx:auth_asym_id>\n'
s+='\t\t\t<PDBx:auth_atom_id>'+atom['atom_id']+'</PDBx:auth_atom_id>\n'
s+='\t\t\t<PDBx:auth_comp_id>'+atom['residue_type']+'</PDBx:auth_comp_id>\n'
s+='\t\t\t<PDBx:auth_seq_id>'+str(atom['residue_pos'])+'</PDBx:auth_seq_id>\n'
s+='\t\t\t<PDBx:group_PDB>ATOM</PDBx:group_PDB>\n'
if atom['alt_id'] is not None:
s+='\t\t\t<PDBx:label_alt_id xsi:nil="true" />\n'
else:
atom['alt_id']='A'
s+='\t\t\t<PDBx:label_asym_id>'+atom['alt_id']+'</PDBx:label_asym_id>\n'
s+='\t\t\t<PDBx:label_atom_id>'+atom['atom_id']+'</PDBx:label_atom_id>\n'
s+='\t\t\t<PDBx:label_comp_id>'+atom['residue_type']+'</PDBx:label_comp_id>\n'
s+='\t\t\t<PDBx:label_entity_id>1</PDBx:label_entity_id>\n'
s+='\t\t\t<PDBx:label_seq_id>'+str(atom['residue_pos'])+'</PDBx:label_seq_id>\n'
s+='\t\t\t<PDBx:occupancy>'+str(atom['occupancy'])+'</PDBx:occupancy>\n'
s+='\t\t\t<PDBx:pdbx_PDB_model_num>'+str(atom['model_no'])+'</PDBx:pdbx_PDB_model_num>\n'
s+='\t\t\t<PDBx:type_symbol>'+atom['atom_element']+'</PDBx:type_symbol>\n'
s+='\t\t</PDBx:atom_site>\n'
s+='\t</PDBx:atom_siteCategory>\n'
s+='</PDBx:datablock>'
return s
Code usage:
mol: RNA_Molecule #suppose a declared instance of RNA_Molecule
rna_io=RNA_IO()
rna_io.write(mol, "7eaf_object.xml",'PDBML')
example output:
cat 7eaf_object.xml
<?xml version="1.0" encoding="UTF-8" ?>
<PDBx:datablock datablockName="7EAF"
xmlns:PDBx="http://pdbml.pdb.org/schema/pdbx-v50.xsd"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://pdbml.pdb.org/schema/pdbx-v50.xsd pdbx-v50.xsd">
<PDBx:atom_siteCategory>
<PDBx:atom_site id="OP3">
<PDBx:B_iso_or_equiv>110.87</PDBx:B_iso_or_equiv>
<PDBx:Cartn_x>-9.698</PDBx:Cartn_x>
<PDBx:Cartn_y>3.426</PDBx:Cartn_y>
<PDBx:Cartn_z>-31.854</PDBx:Cartn_z>
<PDBx:auth_asym_id>A</PDBx:auth_asym_id>
<PDBx:auth_atom_id>OP3</PDBx:auth_atom_id>
<PDBx:auth_comp_id>G</PDBx:auth_comp_id>
<PDBx:auth_seq_id>1</PDBx:auth_seq_id>
<PDBx:group_PDB>ATOM</PDBx:group_PDB>
<PDBx:label_asym_id>A</PDBx:label_asym_id>
<PDBx:label_atom_id>OP3</PDBx:label_atom_id>
<PDBx:label_comp_id>G</PDBx:label_comp_id>
<PDBx:label_entity_id>1</PDBx:label_entity_id>
<PDBx:label_seq_id>1</PDBx:label_seq_id>
<PDBx:occupancy>1.0</PDBx:occupancy>
<PDBx:pdbx_PDB_model_num>1</PDBx:pdbx_PDB_model_num>
<PDBx:type_symbol>O</PDBx:type_symbol>
</PDBx:atom_site>
...
a minor addition to RNA_IO class was made to include the option of writing in PDBML format.
class RNA_IO:
def __init__(self):
...
self.__writers={"PDB": PDB_Writer(),'PDBML': PDBML_Writer(),'XML': PDBML_Writer()}
Parallelism with PDB_Writer
- user interface: ```python rna_io=RNA_IO() mol: RNA_Molecule
rna_io.write(mol, “7eaf_object.xml”,’PDBML’) #also works by specifying XML rna_io.write(mol, “7eaf_object.pdb”,’PDB’)
| PDB_Writer | PDBML_Writer |
|------------|--------------|
| inherits RNA_Writer abstract class | inherits RNA_Writer abstract class |
| `write(molecule: RNA_Molecule, file_path: str)` | `write(molecule: RNA_Molecule, file_path: str)` |
| takes an RNA_Molecule object | takes an RNA_Molecule object |
| uses processor instance to get the atom information | uses processor instance to get the atom information |
| uses processor.flattenMolecule() | uses processor.flattenMolecule_to_dict() |
| has format specific private method `_format_atom_info()` and `_format_molecule_info` | has format specific private method `_format_atom_info()` and `_wrap_str_to_xml()` |
| writes the pdb file | writes the pdbml file |
## Main Implementation using Design Patterns
The implementation of the classes is available in the [src](https://github.com/rna-oop/2425-m1-geniomhe-group-6/tree/main/lab3/src) directory in the `main` branch.
### Demo
For a demonstration of the Builder and Visitor Design Patterns, you can check the notebook [reading-writing.ipynb](/2425-m1-geniomhe-group-6/lab3/demo/reading.ipynb)
[](./demo/reading_writing.ipynb)
### Library Structure
In this lab, we added `Processing` module that contains the classes of the `Builder Design Pattern` and `visitors_writers` module that contains the classes of the `Visitor Design Pattern`inside `IO` module.
The classes are organized in modules and submodules as follows:
```text
.
├── Families
│ ├── __init__.py
│ ├── clan.py
│ ├── family.py
│ ├── species.py
│ └── tree.py
├── IO
│ ├── RNA_IO.py
│ ├── __init__.py
│ ├── parsers
│ │ ├── PDB_Parser.py
│ │ ├── RNA_Parser.py
│ │ ├── __init__.py
│ └── visitor_writers
│ ├── __init__.py
│ ├── pdb_visitor.py
│ ├── visitor.py
│ └── xml_visitor.py
├── Processing
│ ├── ArrayBuilder.py
│ ├── Builder.py
│ ├── Director.py
│ ├── ObjectBuilder.py
│ └── __init__.py
├── Structure
│ ├── Atom.py
│ ├── Chain.py
│ ├── Model.py
│ ├── RNA_Molecule.py
│ ├── Residue.py
│ └── __init__.py
└── utils.py
10 directories, 21 files
Class Diagram
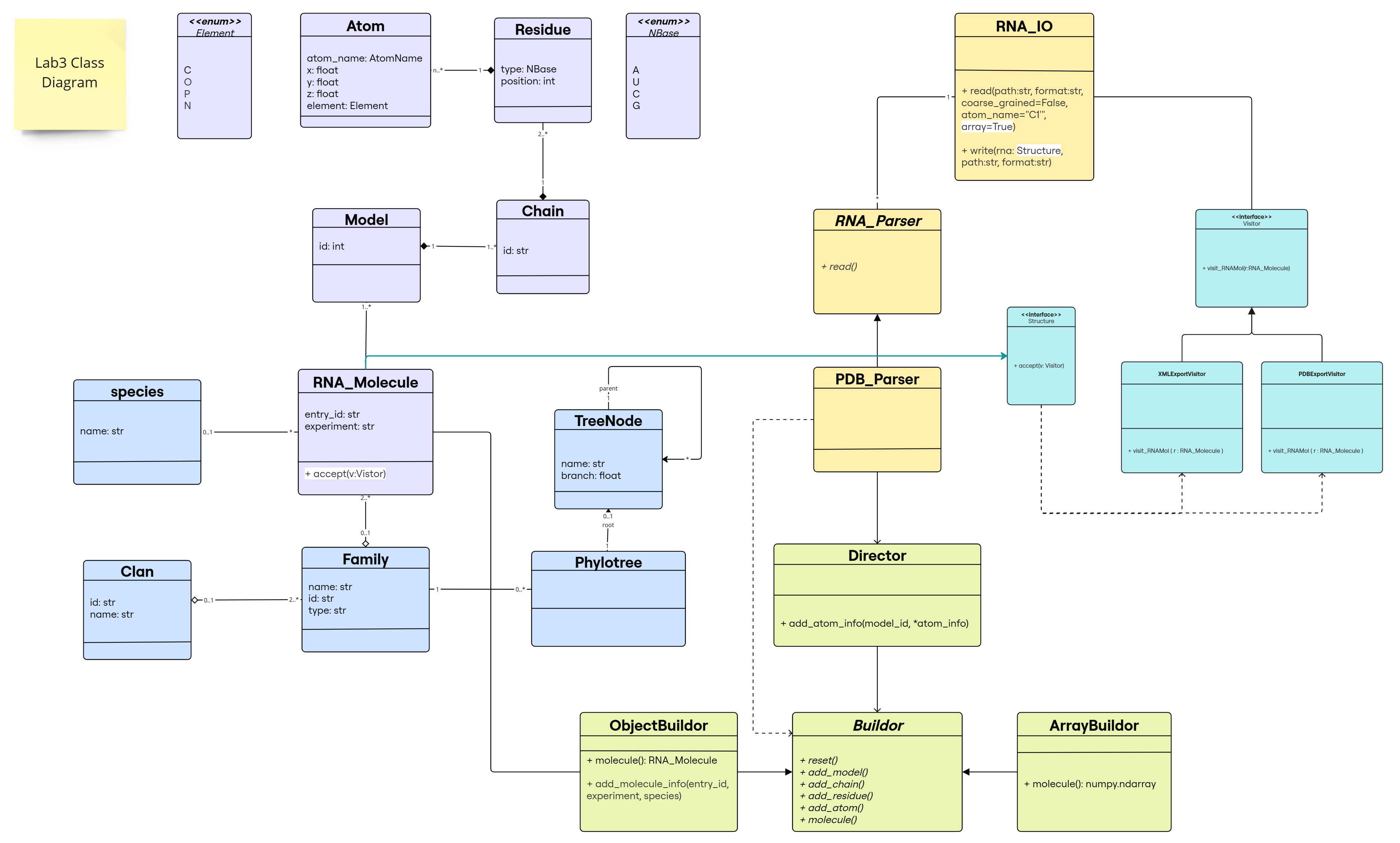
The changes are following this color scheme:
- Builder Design Pattern: light green

- Visitor Design Pattern: light blue
 (classes and inheritance link for our predefined RNA_Molecule class)
(classes and inheritance link for our predefined RNA_Molecule class) - methods added to the existing classes: highlighted in white
Object Diagram
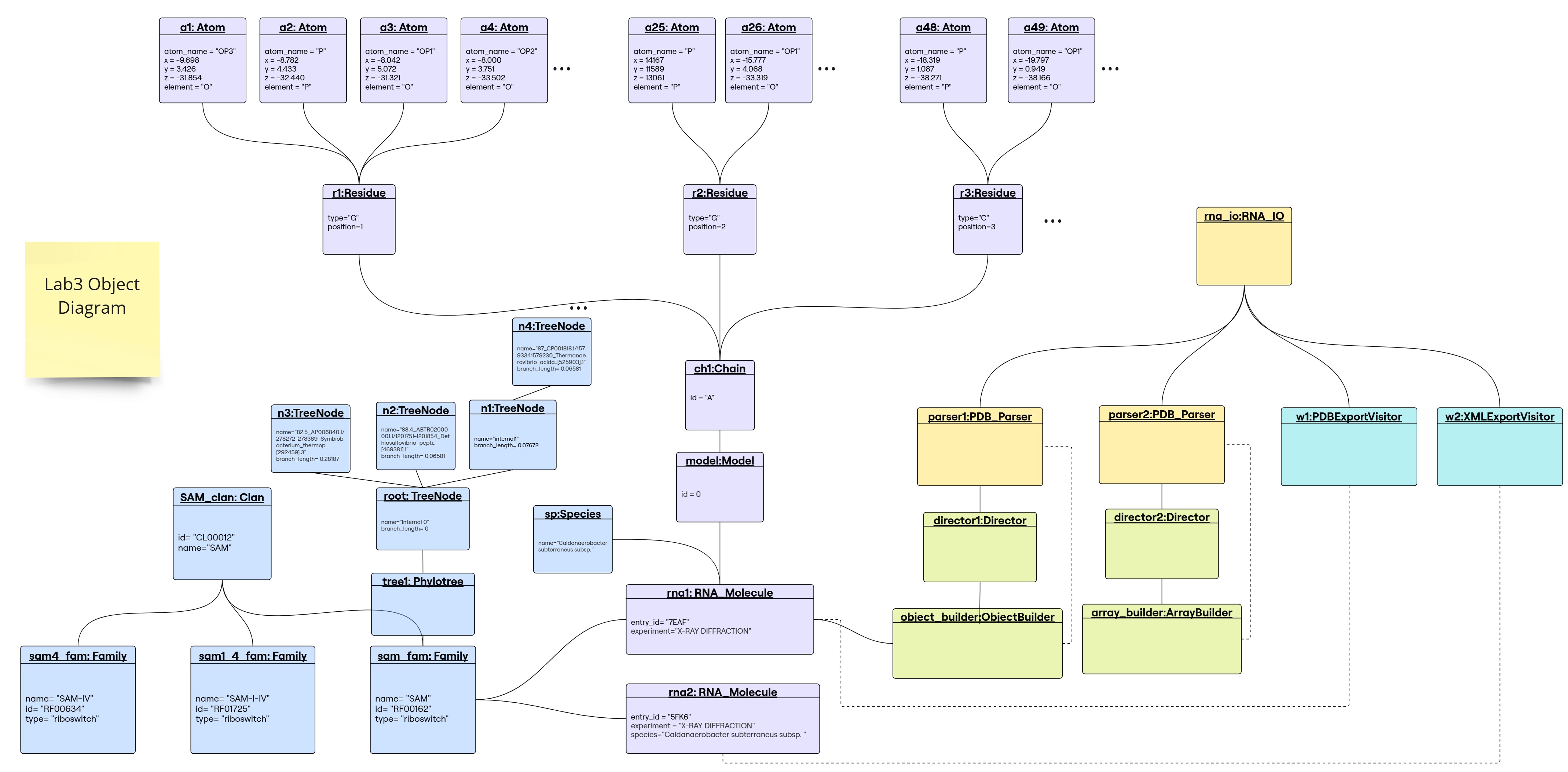
1. Builder Design Pattern
In this lab, we used the Builder design pattern to separate the construction of the RNA molecule object from its representation.
Modifications done to PDB Parser:
-
Added a new argument
array=Trueto theread()function to return the numpy array representation of the molecule. - Instead of using the
Processorclass to create the molecule object when reading the PDB file, we used theDirectorclass and the correspondingBuilderclass.director = Director() if array == True: builder = ArrayBuilder() else: builder = ObjectBuilder() director.builder = builder - Instead of
processor.molecule_info()that used to store the molecule info (id, experiment, species), we added a new method in theObjectBuilderclass calledadd_molecule_info()that adds the molecule info to the molecule object, and we used it in thePDB_Parser’sread() functionas follows:if array==False: builder.add_molecule_info(*molecule_info) -
Instead of
processor.atom_infothat used to store the atom info, we now usedirector.add_atom_info(model_id, *atom_info)which serves as a recipe to build the corresponding molecule. - Finally, instead of
processor.createMolecule()that creates only an object, we returnbuilder.moleculeto get the built molecule that can be an object or a numpy array depending on the builder used.
i. Director class
-
The
Directorclass serves as a director for theBuilderclasses. - Attributes:
__builder: The builder object that will be used to build the object. Initialized toNone.
- Methods:
- @property
builder: Returns the builder object. - @builder.setter
builder: Sets the builder object after checking if it is an instance of theBuilderclass. add_atom_info: serves as a common recipe on how to build the molecule for the differentBuilderclasses:def add_atom_info(self, model_id, *atom_info): atom_name, x, y, z, element, residue_name, residue_id, chain_id, altloc, occupancy, temp_factor, i_code, charge = atom_info self.builder.add_model(model_id) self.builder.add_chain(chain_id) self.builder.add_residue(residue_name, residue_id, i_code) self.builder.add_atom(atom_name, x, y, z, element, altloc, occupancy, temp_factor, charge)- The method takes the model_id and atom_info as arguments that are retrieved from the
Parserand calls the corresponding methods in the builder object in a specific order to add the model, chain, residue, and atom information.
- The method takes the model_id and atom_info as arguments that are retrieved from the
- @property
ii. Builder class
-
The
Builderclass is an interface, implemented as an abstract class with all its methods being abstract. -
It defines the methods that the concrete
Builderclasses should implement.
@property
@abstractmethod
def molecule(self):
pass
@abstractmethod
def add_atom(self):
pass
@abstractmethod
def add_residue(self):
pass
@abstractmethod
def add_chain(self):
pass
@abstractmethod
def add_model(self):
pass
@abstractmethod
def reset(self):
pass
- The
moleculemethod is a property that should return the built molecule object. - The
resetmethod is used to reset the builder object to its initial state. - The other methods are used to add atom, residue, chain, and model information to the molecule object.
iii. ObjectBuilder class
- The
ObjectBuilderclass is a concrete builder class that implements theBuilderinterface. -
It is responsible for constructing the RNA molecule object.
- Attributes:
__molecule: The RNA molecule object that is being built.- other private attributes to keep track of the model, chain, and residue ids.
-
Methods:
reset: Resets the builder object to its initial state:- The molecule object is set to an empty RNA molecule object.
- The model, chain, and residue ids are set to 0.
molecule: @property, returns the built molecule object and resets the builder object.
-
add_model:- Creates a new model object with the given model_id and adds it to the molecule object.
- Sets the model_id attribute to the given model_id.
add_chain:- Retrieves the model object with the current model_id.
- Creates a new chain object with the given chain_id and adds it to the model object.
- Sets the chain_id attribute to the given chain_id.
add_residue:- Retrieves the chain object with the current chain_id from the model object with the current model_id.
- Creates a new residue object with the given residue arguments and adds it to the chain object.
- Sets the residue_id attribute to the residue_id of the added residue.
add_atom:- Retrieves the residue object with the current residue_id from the chain object with the current chain_id from the model object with the current model_id.
- Creates a new atom object with the given atom arguments and adds it to the residue object.
add_molecule_info:- Additional method specific for this builder.
- Adds the entry_id, experiment, and species information to the molecule object.
iv. ArrayBuilder class
-
The
ArrayBuilderclass is a concrete builder class that implements theBuilderinterface. - It is responsible for constructing the numpy array representation of the RNA molecule object:
- Dimension:
(number of models, max_residues_no, max_atoms_per_residue_no, 3) - Stores the x,y,z coordinates of each atom.
- Fills the array with nan values where there is no atom information.
- Dimension:
- Attributes:
__array: A dictionary that stores the atom coordinates for each residue in the molecule.__model_id: The current model id.__residue_id: The current residue id.__prev_atom: A list that stores the previous atom name and occupancy to handle alternate locations.
-
Methods:
reset: Resets the builder object to its initial state:- The array dictionary is set to an empty dictionary.
- The model_id and residue_id are set to 0.
- The prev_atom list is set to empty string for atom_name and 0.0 for occupancy.
add_model: Sets the model_id attribute to the given model_id.add_chain: Does nothing.add_residue: Sets the residue_id attribute to the given residue_id and initializes the atom list for the residue.add_atom: Adds the atom coordinates to the atom list of the current residue in the array dictionary.- Handles alternate locations by storing the atom with the highest occupancy.
molecule: @property, returns the numpy array representation of the molecule.- Finds the maximum number of atoms per residue by taking the maximum length of the atom lists in the array dictionary.
- Finds the maximum number of residues by taking the maximum residue_id from the keys of the array dictionary.
- Creates a numpy array with the calculated dimensions and fills it with the atom coordinates.
- Resets the builder object to its initial state.
- Importance of the
dictionary:- Since we cannot know the maximum dimensions of the numpy array in advance, we used a dictionary to store the atom coordinates for each residue.
- The keys are tuples of the form
(model_id, residue_id)and the values are lists of atom coordinates. - This structure allows for easy retrieval and addition of atom coordinates for each residue.
- The dictionary is cleared after constructing the numpy array.
2. Visitor design pattern
The visitor design pattern aims to separate the algorithm from the object structure on which it operates. In this case we want to be able to operate on RNA to export it into various file formats: PDB and PDBML/XML. Our aim is to perform this without adding a functionality to the RNA_Molecule class itself (through decoupling), but rather to create a new class that will be able to visit the RNA molecule and perform the export functionality.
Modifications done to PDB_Writer
As defined in the lab2, The PDB_Writer class was reposnible for generating the PDB file, we are compare it here directly with PDBExportVisitor class in the visitor design pattern, which has the same responsibility.
The same functionalities writing the file persists, however, only some technicalities of handeling them are changed to account for class and method terminology and manipulation consistency with the way the Visitor pattern in defined. On the other hand, it’s almost exactly identical to the previous implementation (more on that in the next section).
In addition, since the flattening methods in processor are not really instance-related methods, we have moved these to utils (flattenMolecule and flattenMolecuke_to_dict) because it would be interesting to allow the user to use these functions directly if needed (like in torch).
| PDB_Writer | PDBExportVisitor |
|---|---|
implements RNA_Writer interface to enforce a method |
implements Visitor interface to enforce a method |
| no caching | no caching |
write method |
visit_RNA_Molecule method |
takes RNA_Molecule object |
takes RNA_Molecule object |
| generates PDB file | generates PDB file |
uses _format_atom_info method |
uses _format_atom_info method |
uses flattening through flattenMolecule, method of processor (static behavior) |
uses flattening through flattenMolecule_to_dict, added function to utils |
[!NOTE] As can be seen, in our previous decoupled design,
PDB_Writermet an equivalent implementation of theVisitordesign pattern: every row in the table above is a direct correspondence.
Slight differences are within what the Visitor design pattern enforces, which is the visit method that will be called on the RNA_Molecule object, and the accept method that will be called on the RNA_Molecule object to accept the visitor.
This design pattern thus can be used in the following way:
# rna_Molecule: RNA_Molecule
pdb_exporter = PDBExportVisitor()
rna_Molecule.accept(pdb_exporter)
xml_exporter = XMLExportVisitor()
rna_Molecule.accept(xml_exporter)
In order to maintain an efficient user-agnostic interface that keeps export encapsulated, we have maintained the same RNA_IO class, with the same write method, that will take care of the visitor pattern implementation.
First, it declares visitor instances in one of its rna_io object’s attributes:
self.__writers={"PDB": PDBExportVisitor(), "XML": XMLExportVisitor(), "PDBML": XMLExportVisitor()}
def write(self, structure, format):
if format not in self.__writers:
raise ValueError(f"Format {format} is not supported")
exporter=self.__writers[format]
structure.accept(exporter)
Slight intro and background explaining the design:
The visitor design pattern is composed of the following elements:
| entities | type | description |
|---|---|---|
Structure |
interface | defines the accept method that will be implemented by the concrete elements of the object structure |
RNA_Molecule |
class | implements the Structure interface and defines the accept method that will call the visit method of the visitor, our class type defined throughout the project |
Visitor |
interface | enforce a visit method on all concrete visitors that implements it |
PDBExportVisitor |
class | implements Visitor and defines the visit method for each element of the object Structure (here only RNA_Molecule) $\leftarrow$ exports a PDB file |
XMLExportVisitor |
class | implements Visitor and defines the visit method for each element of the object Structure (here only RNA_Molecule) $\leftarrow$ exports a PDBML/XML file |
i. Visitor interface
The visitor interface is defined in the visitor module and contains a single method, visit_RNA_Molecule, which will be implemented by the concrete visitors
# visitor.py
class Visitor(ABC):
@abstractmethod
def visit_RNA_Molecule(self, rna: RNA_Molecule):
pass
There are 2 concrete visitors in this lab, found in submodules pdb_visitor and xml_visitor. Each of these classes implements the Visitor interface and defines the visit_RNA_Molecule method, which will perform the export operation.
ii. PDBExportVisitor class
PDBExportVisitor# pdb_visitor.py class PDBExportVisitor(Visitor): def visit_RNA_Molecule(self, rna: RNA_Molecule): # export the RNA molecule to a PDB file ...this class has some helper methods in order to format the data in the PDB format, which are private methods( start with `_ and not part of the class) not meant to be accessed.
iii. XMLExportVisitor class
XMLExportVisitor# xml_visitor.py class XMLExportVisitor(Visitor): def visit_RNA_Molecule(self, rna: RNA_Molecule): # export the RNA molecule to a PDBML/XML file ...Formatting into XML is done manually without the use of
xmllibrary.
Both classes use flattening functions provided in utils to convert the RNA_Molecule object into a list or a dictionary, which can then be used to generate the file (primitive data types to be used instead of objects, makes it more flexible to implement other similar visit methods for other types of objects).
iv. Structure interface
this is the Component interface, as per the design pattern definition. Different ConcreteComponents classes implement this interface
In this implementation, the Structure interface is implemented by the RNA_Molecule module (because RNA_Molecule will implement it) and defines the accept method that takes any Visitor as an argument and enforces it on ConcreteElement classes that implement it.
Underneath the hood, the accept method of the RNA_Molecule class will call the visit method of the visitor class, which will then perform the export operation.
class RNA_Molecule(Structure):
...
def accept(self, visitor: Visitor):
visitor.visit_RNA_Molecule(self)
[!WARNING] since python does not provide direct overloading, we need to define a method for each element of the object structure that the visitor will visit. In this case, we have the
RNA_Moleculeclass (instead of having severalvisitmethods that take different types of arguments like in java, it’ll bevisit_RNA_Molecule()).
In our case, there is only one ConcreteComponent that implements Structure, which is the RNA_Molecule class, however, this decoupled design allows us at any point to define different concrete Structure classes (e.g. different types of molecules like DNA_Molecule, protein_Molecule, etc. or different RNA mol representation objects like a RNA_ndarray class)
The logic RNA_Molecule will accept any visitor type, which will call the visit method for RN_Molecule of the specific visitor instanciated
graph TD
A[RNA Molecule] -.-> B[Visitor]
B -.-> C[PDB Export Visitor]
B -.-> D[XML Export Visitor]
style A fill:#12387F,stroke:#333,stroke-width:2px
style B fill:#12387F,stroke:#333,stroke-width:2px
style C fill:#12387F,stroke:#333,stroke-width:2px
style D fill:#12387F,stroke:#333,stroke-width:2px
Advantages and Disadvantages
For the Builder Design Pattern
What we might consider as a disadvantage of the Builder Design Pattern over the previous implementation:
- The Builder Design Pattern adds complexity to the code by introducing additional classes and methods.
- The previous model just required a single additional method
createArray()in theProcessorclass to return the numpy array representation of the molecule.
But on the other hand, it has also introduced many advantages:
- Each representation has its own dedicated builder class, making the code cleaner and more maintainable, especially if more representations are needed in the future.
- The Builder Pattern enables the direct construction of the required representation without unnecessary intermediate objects.
- It provides a single recipe for the construction, that is common to all representations, which ensures consistency in how different representations are generated, reducing redundancy and potential errors.
- It breaks the construction into smaller steps, making it easier to modify or extend the building process without affecting the entire system, allowing for greater flexibility and maintainability.
For the Visitor Design Pattern
As previously discussed, equivalent notions have been found between this and the prior implementation (check visitor pattern explanation)
There is 2 advantages of the Visitor Design Pattern, only thanks to its indirect relation to the object structure (accept method that takes a Visitor object):
- The user now can perform exporting from an
RNA_Moleculeobject, a functionality that we wanted to have access to initially while keeping the object clean and decoupled. Noting that it can take any visitor type, it allows for a more flexible and extensible design. - Having a
Structureparent, we can add as many as children as we want in the future without needing to worry about decoupling them everytime we implement them, but rather directly overriding theacceptmethod in the new class and providingVisitorwith an overloadedvisitmethod (efficiency in terms of code writing).
-
PDBML: the representation of archival macromolecular structure data in XML. ↩

